Abstract
Disease monitoring with liquid biopsy would be particularly useful in diffuse large B-cell lymphoma (DLBCL), an aggressive hematologic malignancy with poor outcomes in a significant subset of patients. Exosomes are RNA-containing extracellular vesicles which are released from cells and can bind to or be taken up by other cells with resulting biologic effects. We have previously shown that exosomes can be isolated from DLBCL cell lines and primary samples and traced to the cell of origin (Rutherford et al, ASH 2014). We aimed to determine if DLBCL-derived exosomes, which can be extracted from peripheral blood, share a mutational profile with their cell of origin. We hypothesized that detecting mutations in exosomal RNAs would offer advantage over cfDNA profiling due to higher quantities of exosomal RNAs in circulation, better preservation of packaged RNAs, ability to trace exosomes to their neoplastic cell of origin, and enrichment for driver mutations found in expressed RNAs. We performed exome and total RNA sequencing and mutational calling in 5 DLBCL cell lines, followed by mutational calling using total RNA sequencing of exosomal RNAs isolated from the same cell lines and creating a computational algorithm using exome sequencing data as a gold standard.
RNA-seq reads generated from whole cells and exosomes were used for SNP calling. Sequencing data preprocessing was analyzed using JAX civet pipeline (version 4.3.7). Reads were trimmed based on quality scores and aligned using Tophat (version 2.0.13). We then used picard (version 1.95) for reads sorting and GTAK (version 3.4-0) for variant calling. The snps were annotated with snpEff (version 3.6c). All synonymous mutations were filtered out, followed by filtering of non-synonymous mutations with less than 20% allele frequency. Mutations that passed these filters were compared between the cell of origin and corresponding exosomes. The significance of overlap between cell of origin and exosome mutations was evaluated using hypergeometric test.
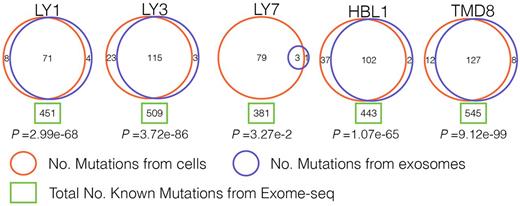
Our approach revealed striking results: exosomal RNAs shared a significant number of mutations found in the cell of origin. In the 5 cell lines almost all mutations, which were expected from exome sequencing analysis, were identified based on analysis of exosomal RNAs (Figure 1; in all five cell lines p<3.27e-2; hypergeometric test). Even though exosomes contain only a small subset of RNA species compared to whole cells, nearly one-third of all mutations detected in the cell exomes were also identified in exosomes, suggesting that mutated RNAs are enriched in exosomes. As an example, OCI-Ly1 cell line carries 451 mutations based on exome analysis and 79 of those are expected to be present in exosomal RNA species based on packaged RNA content; 71 expected and 4 novel were identified through our analysis of OCI-Ly1 exosomes.
Given that exosomes circulate through the body and can be easily procured from peripheral blood, we anticipate that analysis of RNA in exosomes could provide important information for disease monitoring and therapeutic decision making in DLBCL. This approach is advantageous compared to analysis of circulating tumor cell or cfDNA because exosomal RNA content is well preserved due to the vesicular structure, exosomes can be delineated based on the cell of origin, and they contain expressed RNA species, giving clues about aberrant transcriptional programming of the tumor, which can then be targeted. We also showed that exosomes seem to preferentially package RNAs that carry mutations, and therefore may serve yet an unknown function. That feature also allows sampling mutated RNAs that are expressed in the tumor with many likely coding for neoantigens. If this finding can be validated in larger studies, it may enable targeted profiling of tumor neoantigens using liquid biopsy. We are actively designing clinical trials in which we will monitor the content of exosomes in the peripheral blood of patients with DLBCL prior to, during and following treatment with standard and novel agents. This will provide us with insight into changes in exosomal content in those patients with optimal and suboptimal responses to treatment, so that we can design more effective therapeutic strategies in those who are not cured with standard chemoimmunotherapy.
Rutherford: Juno Therapeutics Inc.: Consultancy, Honoraria; Genentech: Research Funding. Li: The Jackson Laboratory for Genomic Medicine: Employment. Jiang: Genentech: Employment.
Author notes
Asterisk with author names denotes non-ASH members.

This feature is available to Subscribers Only
Sign In or Create an Account Close Modal